The PG-URE estimator is a statistical tool that can be used to assess the performance of image denoising algorithms in presence of mixed Poisson-Gaussian noise. It was introduced in the following publication:
Yoann Le Montagner, Elsa Angelini, and Jean-Christophe Olivo-Marin, An unbiased risk estimator for image denoising in the presence of mixed Poisson-Gaussian noise, IEEE Transactions on Image Processing 23(3), pp. 1255–1268, 2014.
This article is freely available on IEEE Xplore (Open Access paper).
Matlab code
In the spirit of reproducible research, the Matlab code and the data used to generate the results presented in this publication is provided here: [ZIP].
A “README” file is provided, which gives a brief description of the package content and of the installation procedure. In particular, to work properly, this package requires:
- The standard Matlab Wavelet tool-box.
- The NESTA optimization tool-box, available freely at http://statweb.stanford.edu/~candes/nesta/.
- The NL-means tool-box, available freely at http://www.mathworks.com/matlabcentral/fileexchange/13619-toolbox-non-local-means.
All publications describing research work using this code must cite the publication mentioned above.
Authors
Yoann Le Montagner (yoann.le-montagner -at- m4x.org)
Elsa Angelini (elsa.angelini -at- telecom-paristech.fr)
Jean-Christophe Olivo-Marin (jcolivo -at- pasteur.fr)
Automatic quantification of immunofluorescence images relies either on the detection and counting of spots superimposed on biological structures, usually immersed in a non-uniform background, or on the outlining of larger cellular compartments. We have developed methods for spot detection and characterization that allow a fast and reproducible quantitative analysis of these images. based on a multiscale approach that uses a shift invariant discrete wavelet transform(SI-DWT) and on the selective filtering of wavelet coefficients. This scheme allows to separate and characterize objects of different sizes by selecting only a vicinity of detail images with corresponding scales adapted to the size of the spots. The extraction step consists in retaining the significant responses of the locally supported detail signal filters to the desired features, at the different scales of the wavelet representation. This is accomplished through a denoising technique using a threshold value which is image and level dependent and which can be computed automatically from the data. The program detects spots both in 2D and in 3D images and is used by several biological scientists to quantify their images.
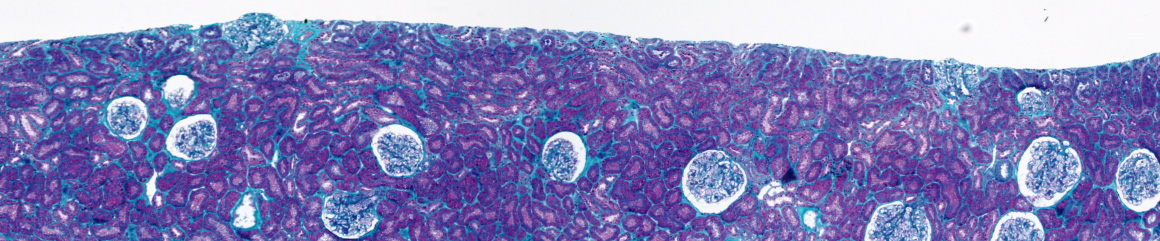
To characterize histopathological images stained by different colorations, we have developed a system to analyse color images. Images are segmented by a split and merge approach and by color quantization, to reduce color classes. We have also defined a criterion to choose the best color space. This method has been applied successfully to the quantification of interstitial fibrosis quantification in chronic allograft nephropathy of renal biopsy. The proportion of green to total pixels in the biopsy was then calculated and used as an index of interstitial fibrosis. The results are correlated to the values of quantification realized by an expert.
We have developed a method to perform the detection and the tracking of microscopic spots directly on four dimensional (3D+t) image data. It extends our previous work by being able to detect with high accuracy multiple biological objects moving in three-dimensional space and by incorporating the possibility to follow moving spots switching between different dynamics characteristics. Our method is based on a two step procedure: first, the objects are detected in the image stacks thanks to a procedure based on a three-dimensional wavelet transform; then the tracking is performed within a Bayesian framework where each object is represented by a state vector evolving according to biologically realistic dynamic models. The main advantage of wavelet-based detection is to be robust to the local variation of contrast and to the imaging noise. The Bayesian tracking allows to predict the new position of a spot knowing its past positions and increases the reliability of the data association step.
Accurate computational localization of single fluorescent particles is of interest to many biophysical studies and underlies recent approaches to high resolution microscopy using photo-switchable fluorophores. The position of individual particles is typically computed by least-squares fitting of a gaussian intensity profile to the image, whose band-width is either derived from an idealized theoretical model of the point spread function (PSF), or itself fitted to the image. However, the band-width best approximating the actual PSF may differ significantly from its theoretical value, while fitting it is expected to degrade localization accuracy. Here, instead, we measure the real PSF bandwidth using fluorescent beads as calibration probes, and use this new bandwidth in a Gaussian model fitting algorithm. We show on simulated and real images that this simple modification of the standard localization procedure results in significant improvement of the 3D accuracy in the nanometer range.
Keywords: 3D biological imaging, multiple cell tracking, active contours
Summary: Elucidating the mechanisms of cell deformation and motility is a topic of major interest in cell biology, for they are strongly involved in cell development, immune responses, cancer and infectious diseases. Our aim to develop robust and fully automated image analysis tools, able to extract quantitative measures of cell dynamics from multi-dimensional (2D/3D), multi-modal (brightfield/phase-contrast/fluorescence) time-lapse microscopy data. It is nowadays a growing consensus that the combination of large-scale cell imaging and sophisticated computational analysis tools is necessary to provide new insights on numerous biological phenomena. Yet, many studies related to cell shape and motility rely still today on manual (though generally computer-assisted) analysis in 2D sequences. This process is cumbersome and prone to user bias and reproducibility issues. Migration to full 3D time-lapse experiments is a necessary step to reliably quantify cell shape information, but induces new challenges in terms of image acquisition and quantification, yielding a need for more robust and fully automated analysis techniques.
To this end, we have been focusing on the past years upon “active contours” techniques. The principle is to deform an initial contour placed on the image until it fits the boundary of the target cell. The deformation can be mathematically expressed as the minimization of an energy functional, which comprises several terms related either to the image data (driving the contour toward the cell boundary) or to geometrical properties of the contour (regularizing the deformation to avoid local energy minima). This energy is an essential ingredient of the method, and several of our contributions consist in adapting this energy and its implementation to meet the particular constraints of biological imaging, e.g., low signal-to-noise ratio, multiple cell-cell contacts over time, inhomogeneous cell staining, photo-bleaching, etc.. Active contours also offer a flexible formalism for efficient shape representation and quantification, allowing to compute robust statistics over large-scale cell populations.
The algorithms developed for this project are currently used through collaborations for numerous applications including multi-cell segmentation, tracking, interaction and particle localization.