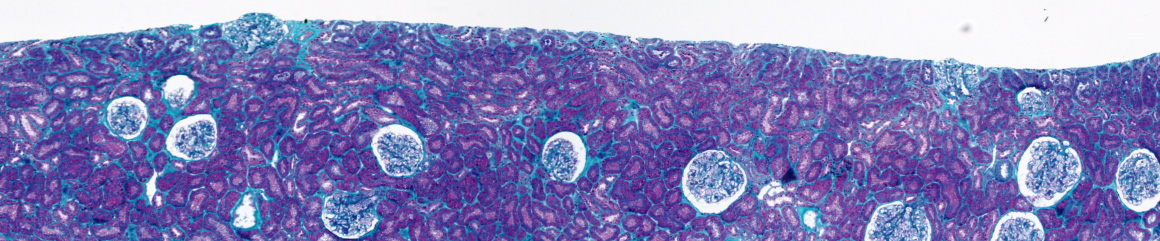
Topological analysis of tissues
Topological analysis of tissues consists in detecting and characterizing the biological structures constituting the tissue architecture. From a medical point of view, it allows the representation of the visual criteria used by physicians to distinguish the grades of disease.
For several years, the topological characterization of the image content is a central tool for several applications. Topological descriptors allow the definition of efficient and, above all, interpretable methods for clustering or classification.
Justifying the model prediction by arguments referring to the tissue morphology improves both the understanding of the studied phenomena and the physician’s confidence. Finally, robust detection of the tissue architecture allows the definition of image-based biomarkers to study the evolution of a disease.
Nowadays, a large literature exists about methods considering topological constraints and their application to microscopy or histopathology [3,5,6].
Most of these works are based on the theory of persistent homology allowing us to define image signatures encoding the topological and geometrical structure of the tissue [1,2,4,7,8].
This can be particularly useful, when exploring new large datasets, to detect different subgroups of tissue architectures. Another application could be the discovery of intermediate grades for a given disease.
The topological approach could help to encode the tissue architecture independently of its acquisition conditions (orientation, artifacts) facilitating a semantic representation of the imaged tissue.
The method will be applied to different datasets of histology, cytology, or microscopy images.
2. Xiaoling Hu, Fuxin Li, Dimitris Samaras, and Chao Chen. Topology-preserving deep image segmentation. In H. Wallach, H. Larochelle, A. Beygelzimer, F. d'Alché-Buc, E. Fox, and R. Garnett, editors, Advances in Neural Information Processing Systems, volume 32. Curran Associates, Inc., 2019.
3. Peter Lawson, Andrew Sholl, J. Brown, Brittany Fasy, and Carola Wenk. Persistent homology for the quantitative evaluation of architectural features in prostate cancer histology. Scientific Reports, 9:1139, 02 2019.
4. Michael Moor, Max Horn, Bastian Rieck, and Karsten Borgwardt. Topological autoencoders. In Hal Daumé III and Aarti Singh, editors, Proceedings of the 37th International Conference on Machine Learning, volume 119 of Proceedings of Machine Learning Research, pages 7045–7054. PMLR, 13–18 Jul 2020.
5. Talha Qaiser, Yee Tsang, Daiki Taniyama, Naoya Sakamoto, Kazuaki Nakane, David Epstein, and Nasir Rajpoot. Fast and accurate tumor segmentation of histology images using persistent homology and deep convolutional features. Medical Image Analysis, 55, 04 2019.
6. Lucille Quénéhervé, Grégoire David, Arnaud Bourreille, Jean-Benoit Hardouin, Gabriel Rahmi, N. Neunlist, Jeremy Bregeon, and Emmanuel Coron. Quantitative assessment of mucosal architecture using computer-based analysis of confocal laser endomicroscopy in inflammatory bowel diseases. Gastrointestinal Endoscopy, 89, 08 2018.
7. Fan Wang, Huidong Liu, Dimitris Samaras, and Chao Chen. Topogan: A topology-aware generative adversarial network. In Andrea Vedaldi, Horst Bischof, Thomas Brox, and Jan-Michael Frahm, editors, Computer Vision – ECCV 2020, pages 118–136, Cham, 2020. Springer International Publishing.
8. Jiaqi Yang, Xiaoling Hu, Chao Chen, and Chialing Tsai. A topological-attention ConvLSTM network and its application to EM images. In Marleen de Bruijne, Philippe C. Cattin, Stéphane Cotin, Nicolas Padoy, Stefanie Speidel, Yefeng Zheng, and Caroline Essert, editors, Medical Image Computing and Computer Assisted Intervention – MICCAI 2021, pages 217–228, Cham, 2021. Springer International Publishing.